ss_site_model <- clarkia_hz |>
mutate(grand_mean = mean(admix_proportion))|>
group_by(site)|>
mutate(y_hat = mean(admix_proportion))|>
ungroup()|>
summarise( ss_model = sum((y_hat - grand_mean)^2),
ss_error = sum((admix_proportion - y_hat)^2),
ss_total = sum((admix_proportion - grand_mean)^2),
r2 = ss_model / ss_total,
n = n(),
n_groups = n_distinct(site))• 19. ANOVA Example
Motivating Scenario: You’re ready to run an ANOVA with more than two groups!
Learning Goals: By the end of this subchapter, you should be able to:
Calculate sums of squares, mean squares, the F statistic, and the associated p-value “by hand.”
Generate and Interpret ANOVA output from R (aov() or lm() pipelines) and relate it to the variance partitioning.
We previously saw how to partition variance components to calculate the relevant sums of squares needed to conduct an ANOVA and estimate the proportion of variance explained by our model. Recall that we found:
Sum of Squares Model The sum of the squared difference between each individual’s predicted value and the grand mean.
Sum of Squares Error The sum of the squared difference between each observed value and their predicted value.
Together these sum to equal
- Sum of Squares Total The sum of the squared difference between each observed value and the grand mean.
Code
library(broom)
library(patchwork)
library(tidyverse)
# ---- Load & prepare data ----
clarkia_hz <- clarkia_hz |>
mutate(tmp = as.numeric(factor(site)) + admix_proportion,
id = factor(id),
id = fct_reorder(id, tmp))
# Fit model
site_lm <- lm(admix_proportion ~ site, data = clarkia_hz)
# Add augment info
plot_dat <- augment(site_lm) |>
mutate(id = row_number(),
mean_admix = mean(admix_proportion, na.rm = TRUE)) |>
mutate(tmp = as.numeric(factor(site)) + admix_proportion,
id = factor(id),
id = fct_reorder(id, tmp))
# Data for mean lines per group
mean_lines <- plot_dat |>
group_by(site) |>
summarise(xstart = min(as.numeric(id)),
xend = max(as.numeric(id)),
mean_val = mean(admix_proportion),
.groups = "drop")
# --- (C) Total deviation ---
c <- ggplot(plot_dat,
aes(x = as.numeric(id), y = admix_proportion, color = site)) +
geom_point(alpha = 0.7, size = 2) +
geom_hline(aes(yintercept = mean_admix)) +
labs(title = " (C) Total deviation",
color = "Site",
y = "Admixture proportion") +
geom_segment(aes(xend = as.numeric(id),
yend = mean_admix),
color = "black", alpha = 0.5, linewidth = 0.5) +
theme_light(base_size = 13) +
theme(axis.title.x = element_blank(),
axis.text.x = element_blank(),
axis.ticks.x = element_blank())
# --- (A) Model deviation ---
a <- ggplot(plot_dat,
aes(x = as.numeric(id), y = admix_proportion, color = site)) +
geom_point(alpha = 0.7, size = 2) +
geom_hline(aes(yintercept = mean_admix)) +
# deviation lines
geom_segment(aes(xend = as.numeric(id), y = .fitted, yend = mean_admix),
color = "black", alpha = 0.5, linewidth = 0.5) +
# fitted line
geom_line(aes(y = .fitted), linewidth = 1.2, show.legend = FALSE) +
# horizontal group means
geom_segment(data = mean_lines,
aes(x = xstart, xend = xend,
y = mean_val, yend = mean_val,
color = site), show.legend = FALSE,
linewidth = 1.2, inherit.aes = FALSE) +
labs(title = "(A) Model deviation +",
color = "Site",
y = "Admixture proportion") +
theme_light(base_size = 13) +
theme(axis.title.x = element_blank(),
axis.text.x = element_blank(),
axis.ticks.x = element_blank())
# --- (B) Error deviation ---
b <- ggplot(plot_dat,
aes(x = as.numeric(id), y = admix_proportion, color = site)) +
geom_point(alpha = 0.7, size = 2) +
geom_hline(aes(yintercept = mean_admix)) +
# deviation lines to fitted
geom_segment(aes(xend = as.numeric(id), yend = .fitted),
color = "black", alpha = 0.5, linewidth = 0.5) +
# fitted line
geom_line(aes(y = .fitted), linewidth = 1.2, show.legend = FALSE) +
# horizontal group means
geom_segment(data = mean_lines,
aes(x = xstart, xend = xend,
y = mean_val, yend = mean_val,
color = site),
linewidth = 1.2, inherit.aes = FALSE, show.legend = FALSE) +
labs(title = "(B) Error (residual) deviation =",
color = "Site",
y = "Admixture proportion") +
theme_light(base_size = 13) +
theme(axis.title.x = element_blank(),
axis.text.x = element_blank(),
axis.ticks.x = element_blank())
# Combine panels with shared y-axis and legend
(a + b + c) +
plot_layout(guides = "collect",
axis_titles = "collect",
axes = "collect_y") &
theme(legend.position = "right",
legend.text = element_text(size = 20),
legend.title = element_text(size = 20),)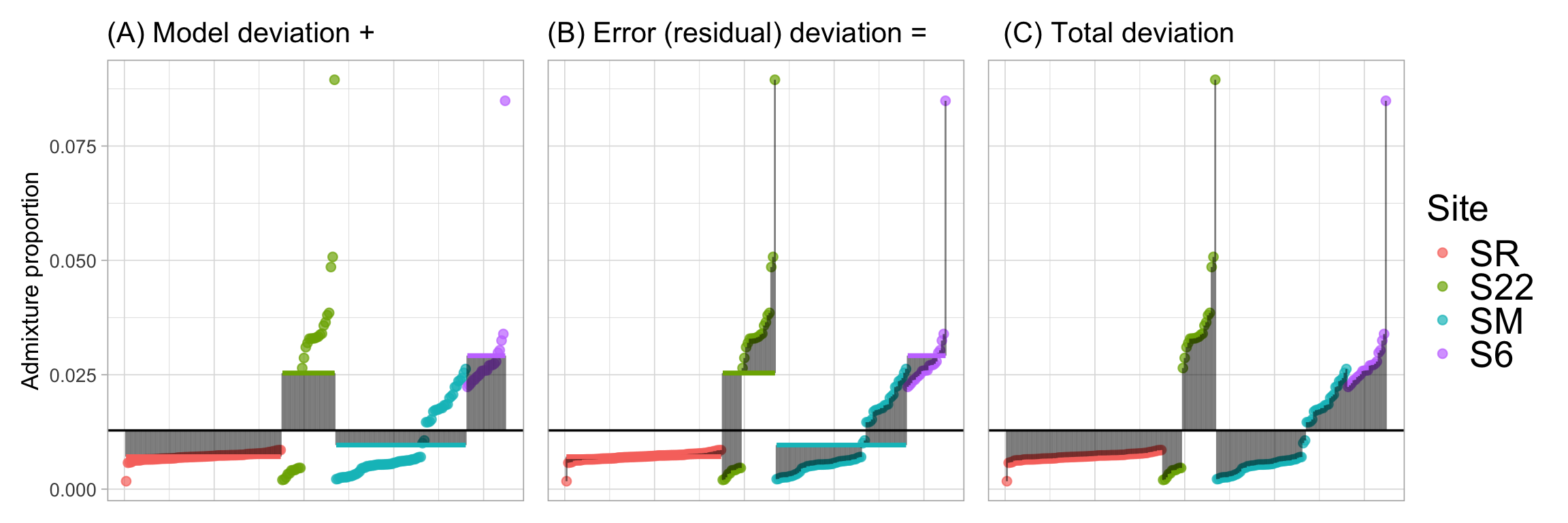
We can use the logic illustrated in Figure 1 to calculate the relevant sums of squares, and the proportion variance explained by the model, \(r^2\):
| ss_model | ss_error | ss_total | r2 | n | n_groups |
|---|---|---|---|---|---|
| 0.0142 | 0.0189 | 0.0331 | 0.4298 | 212 | 4 |
Knowing that \(\text{DF}_\text{model} = n_\text{groups}-1\), and \(\text{DF}_\text{error} = n- n_\text{groups}\), we can find mean squares, \(F\) and our p-value:
pf() looks up our F alue on the F distribution with df1 = df_groups, and df_2 = df_error. Recall that F is a one tailed test – we only want the upper tail (lower.tail = FALSE)
anova_results <- ss_site_model |>
mutate(ms_model = ss_model / (n_groups - 1),
ms_error = ss_error / (n - n_groups),
F_value = ms_model / ms_error,
p_value = pf(F_value , n_groups - 1, n - n_groups, lower.tail = FALSE))| ms_model | ms_error | F_value | p_value |
|---|---|---|---|
| 0.0047379 | 0.0000906 | 52.26634 | 3.2 x 10^-25 |
ANOVA in R
Of course R can do these calculations for us, with either the aov() |> summary() pipeline, or the lm() |> anova() pipeline. both approaches yield the same result as our manual calculation above.
aov() |> summary()
aov(admix_proportion ~ site, data = clarkia_hz)Call:
aov(formula = admix_proportion ~ site, data = clarkia_hz)
Terms:
site Residuals
Sum of Squares 0.01421379 0.01885515
Deg. of Freedom 3 208
Residual standard error: 0.009521017
Estimated effects may be unbalancedaov(admix_proportion ~ site, data = clarkia_hz)|>
summary() Df Sum Sq Mean Sq F value Pr(>F)
site 3 0.01421 0.004738 52.27 <2e-16 ***
Residuals 208 0.01886 0.000091
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1lm() |> anova()
lm(admix_proportion ~ site, data = clarkia_hz)
Call:
lm(formula = admix_proportion ~ site, data = clarkia_hz)
Coefficients:
(Intercept) siteS22 siteSM siteS6
0.007094 0.018282 0.002536 0.022087 lm(admix_proportion ~ site, data = clarkia_hz)|>
anova()Analysis of Variance Table
Response: admix_proportion
Df Sum Sq Mean Sq F value Pr(>F)
site 3 0.014214 0.0047379 52.266 < 2.2e-16 ***
Residuals 208 0.018855 0.0000906
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1