frog_summary <- frogs |>
group_by(treatment) |>
summarise( mean_hatched = mean(hatched.eggs),
sd_hatched = sd(hatched.eggs))• 14. The frogs
Motivating Scenario: You want to know about our data we will use to learn about permutation. You also wouldn’t mind a little refresher on sampling, uncertainty, bootstrapping, and null hypothesis significance testing.
Learning Goals: By the end of this section, you should:
Know enough about the biology and data set presented to get ready for the next sections
Remember how to specify a null and alternative hypothesis
Remember how we use the infer package to
- Calculate a test statistic
- Bootstrap
- Estimate uncertainty
- Calculate a test statistic
Case study: Mate choice & fitness in frogs
There are plenty of reasons to choose your partner carefully. In much of the biological world a key reason is “evolutionary fitness” – presumably organisms evolve to choose mates that will help them make more (or healthier) children. This could, for example explain Kermit’s resistance in one of the more complex love stories of our time, as frogs and pigs are unlikely to make healthy children.
In fact they will form fewer hybrids by mating than will parviflora and xantiana.
To evaluate this this idea Swierk & Langkilde (2019), identified a males top choice out of two female wood frogs and then had them mate with the preferred or nonpreferred female and counted the number of hatched eggs. The raw data are available here

Concept check
It’s an experimental study because the researchers actively intervened and manipulated a variable. They assigned males to mate with either a preferred or a non-preferred female, rather than just observing which mates they chose on their own.
Which of the following describes the biological hypothesis?
The core biological idea is that mate choice should be adaptive. Therefore, the hypothesis is that a male’s choice should lead to a better evolutionary outcome, in this case, higher reproductive success measured by the number of hatched eggs.
- Why not “That there will be a difference in the number of hatched eggs between the two groups”? A biological hypothesis is based on a theory, and therefore predicts a specific direction, such as higher fitness from mate choice. The statement “that there will be a difference” is a non-directional statistical hypothesis; it lacks the underlying biological reasoning and direction.
What is the statistical null hypothesis (\(H_0\))?
The null hypothesis (\(H_0\)) is the hypothesis of ‘no effect.’ It posits that there is no true difference in the average number of hatched eggs between the preferred and non-preferred groups in the population. Any difference we see in our sample is assumed to be due to random sampling variation.
- Why not “The observed difference in mean hatched eggs in our sample is zero.”? Because null hypotheses are about population parameters not observed sample estimates.
Which of the following is a potential source of bias in this experiment?
Bias occurs when there is a systematic difference between your treatment groups that is not the treatment itself. If the non-preferred females were also different in another key aspect (like health or size), we wouldn’t be able to tell if the results were due to ‘preference’ or that other factor. Random assignment helps to prevent this.
Visualize Patterns
Loading, formating, and plotting the data
frogs <- read_csv("https://raw.githubusercontent.com/ybrandvain/biostat/master/data/Swierk_Langkilde_BEHECO_1.csv")|>
dplyr::select(year, pond, treatment,hatched.eggs, total.eggs, num.metamorphosed,field.survival.num)%>%
mutate_at(.vars = c("hatched.eggs", "total.eggs", "num.metamorphosed" , "field.survival.num"), as.numeric)
library(ggforce)
ggplot(frogs, aes(x = treatment, y = hatched.eggs, color = treatment, shape = treatment)) +
geom_violin(show.legend = FALSE)+
geom_sina(show.legend = FALSE, size = 6, alpha = .6) + # if you don't have ggforce, use geom_jitter
stat_summary(fun.data = mean_cl_boot, geom = "pointrange", color = "black", show.legend = FALSE,linewidth = 1.2)+
theme(axis.title = element_text(size = 30),
axis.text = element_text(size = 30))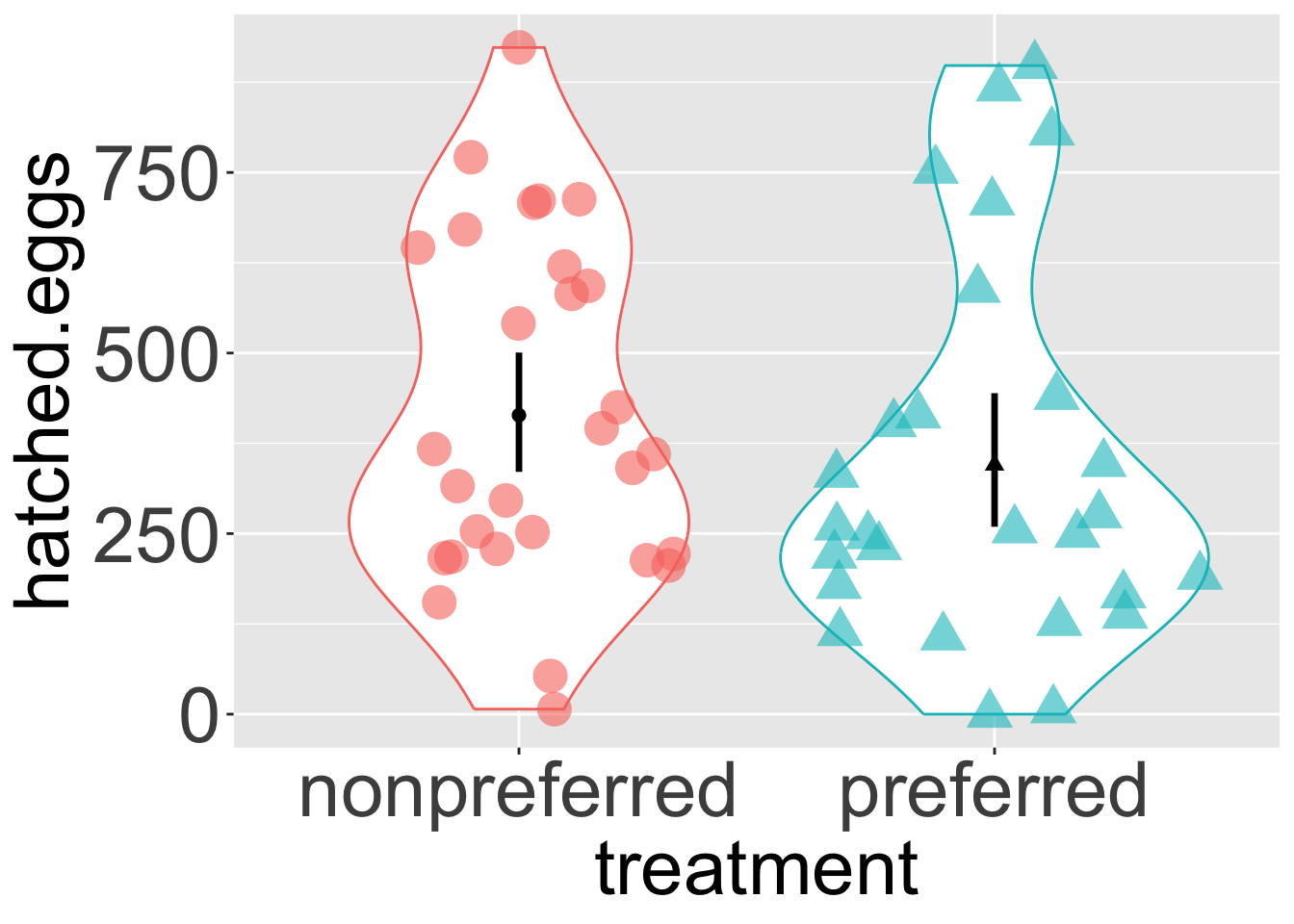
One of the first things we do with a new dataset is visualize it! A quick exploratory plot (refer back to our Intro to ggplot) helps us understand the distribution and shape of our data, and informs us how to develop an appropriate analysis plan.
Estimate Parameters
Now that we have a better understanding of the data’s shape, we can create meaningful summaries by estimating appropriate parameters (see the chapters on data summaries, associations, and linear models).
For the frog mating data, let’s first calculate the mean and standard deviation of the number of hatched eggs for both the preferred and non-preferred mating groups.
| treatment | mean_hatched | sd_hatched |
|---|---|---|
| nonpreferred | 413.9310 | 236.8918 |
| preferred | 345.1111 | 259.6380 |
Calculating a test statistic
How do we summarize this variation down to a single test statistic? A high-level summary could be the difference in mean hatched eggs between the preferred and non-preferred mating groups. We could find this as 345.11 - 413.93 = -68.82. But I want to remind you how to do this with the infer package as we will be using it throughout this section:
library(infer)
frogs |>
specify(hatched.eggs ~ treatment) |>
calculate(stat = "diff in means", order = c("preferred","nonpreferred"))Response: hatched.eggs (numeric)
Explanatory: treatment (factor)
# A tibble: 1 × 1
stat
<dbl>
1 -68.8… So both visual and numerical summaries suggest that, contrary to our expectations, more eggs hatch when males mate with non-preferred females. However, we know that sample estimates will differ from population parameters by chance (i.e., sampling error). So we …
Bootstrap to estimate the difference in means
We can separately bootstrap to estimate the uncertainty in each mean, and then bootstrap the data together to estimate the uncertainty in the difference in means:
Bootstrapping the data
library(infer)
ci_preffered <- frogs |>
filter(treatment == "preferred")|>
specify(response = hatched.eggs) |>
generate(reps = 5000, type = "bootstrap") |>
calculate(stat = "mean")|>
get_ci()
ci_nonpreffered <- frogs |>
filter(treatment == "nonpreferred")|>
specify(response = hatched.eggs) |>
generate(reps = 5000, type = "bootstrap") |>
calculate(stat = "mean")|>
get_ci()
ci_diff <- frogs |>
specify(hatched.eggs ~ treatment) |>
generate(reps = 5000, type = "bootstrap") |>
calculate(stat = "diff in means", order = c("preferred","nonpreferred"))|>
get_ci()
kable(bind_rows(ci_preffered,ci_nonpreffered,ci_diff)|>
mutate_all(round)|>
mutate(type = c("preferred","nonpreferred","difference in means")))| lower_ci | upper_ci | type |
|---|---|---|
| 255 | 441 | preferred |
| 333 | 499 | nonpreferred |
| -194 | 63 | difference in means |
There is considerable uncertainty in our estimates. Our bootstrapped 95% confidence interval includes cases where preferred matings yield more hatched eggs than nonpreferred matings, as well as cases where nonpreferred matings result in more hatched eggs.
How do we test the null hypothesis that mate preference does not affect egg hatching? Let’s generate a sampling distribution under the null!
